Exporting Data¶
Data science is not effective without saving results.
- Another wise person
Applied Review¶
Data in Python¶
- Data is frequently represented inside a DataFrame - a class from the pandas library
- Other structures exist, too - dicts, models, etc.
- Data is stored in memory - this makes it relatively quickly accessible
- Data is session-specific, so quitting Python (i.e shutting down JupyterLab) removes the data from memory
Importing Data¶
- Tabular data can be imported into DataFrames using the
pd.read_csv()function - there are parameters for different options and otherpd.read_xxx()functions.
- Other data formats like JSON (key-value pairs) and Pickle (native Python) can be imported using the
withstatement and respective functions:- JSON files use the
load()function from thejsonlibrary - Pickle files use the
load()function from thepicklelibrary
- JSON files use the
General Model¶
General Framework¶
A general way to conceptualize data export from Python to Disk:
- Data sits in memory in the Python session
- Python code can be used to copy the data from Python's memory to an appropriate format on disk
This framework can be visualized below:
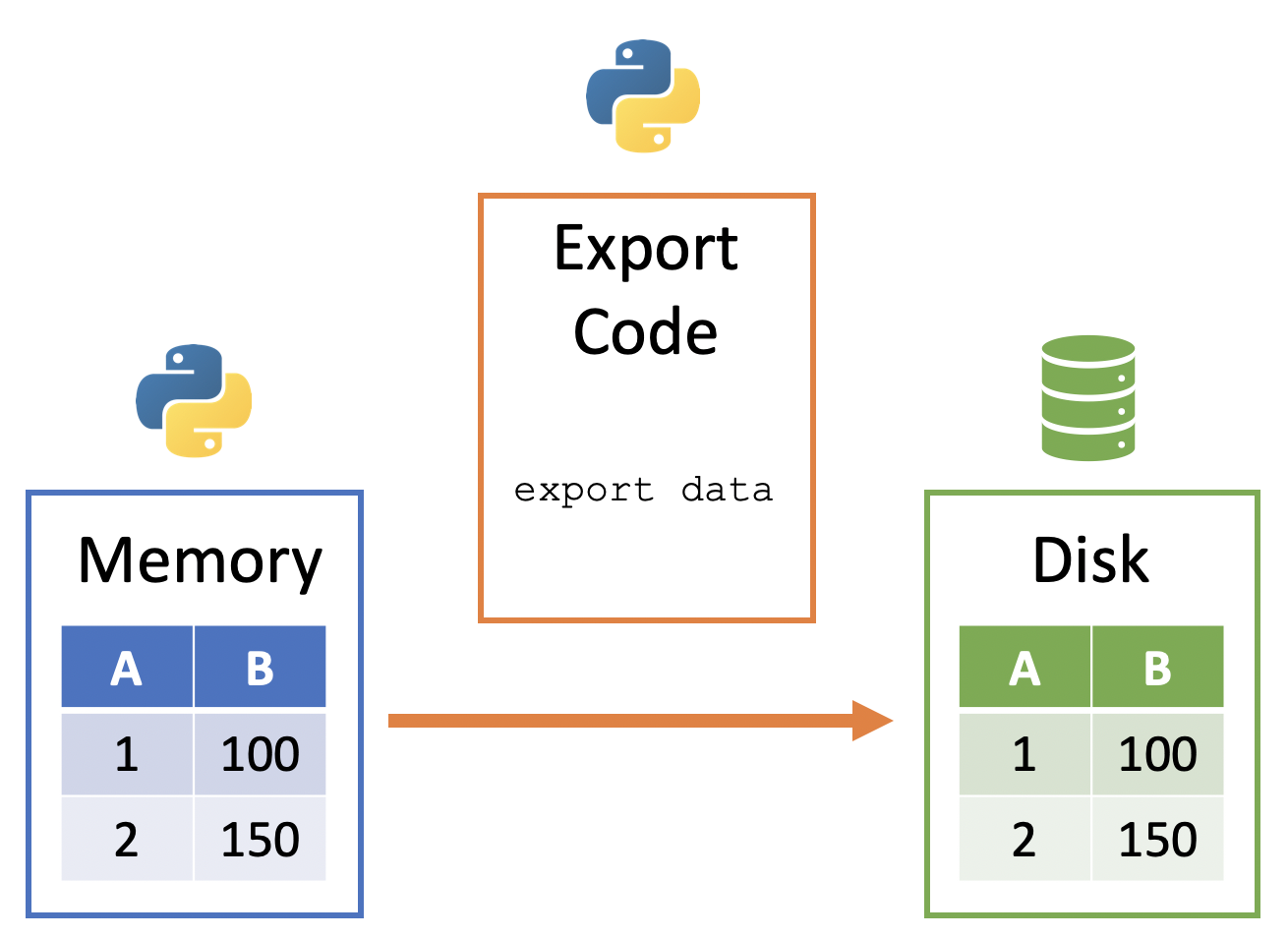
Exporting DataFrames¶
Remember that DataFrames are representations of tabular data -- therefore, knowing how to export DataFrames to tabular data files is important.
Exporting Setup¶
We need data to export.
Let's begin by revisiting the importing of tabular data into a DataFrame:
import pandas as pd
planes_df = pd.read_csv('../data/planes.csv')
Next, let's do some manipulations on planes_df.
Question
How do we select the year and manufacturer variables while returning a DataFrame?
planes_df = planes_df[['year', 'manufacturer']]
Question
How do we compute the average year by manufacturer?
avg_year_by_man_df = (
planes_df.groupby('manufacturer', as_index = False)
.mean()
)
Let's view our result to find the manufacturers with the oldest planes:
avg_year_by_man_df.sort_values('year').head()
| manufacturer | year | |
|---|---|---|
| 16 | DOUGLAS | 1956.000000 |
| 15 | DEHAVILLAND | 1959.000000 |
| 7 | BEECH | 1969.500000 |
| 13 | CESSNA | 1972.444444 |
| 12 | CANADAIR LTD | 1974.000000 |
Exporting DataFrames with Pandas¶
DataFrames can be exported using a method built-in to the DataFrame object itself: DataFrame.to_csv().
avg_year_by_man_df.to_csv('../data/avg_year_by_man.csv')
Let's reimport to see the tabular data we just exported:
pd.read_csv('../data/avg_year_by_man.csv').head()
| Unnamed: 0 | manufacturer | year | |
|---|---|---|---|
| 0 | 0 | AGUSTA SPA | 2001.000000 |
| 1 | 1 | AIRBUS | 2007.201220 |
| 2 | 2 | AIRBUS INDUSTRIE | 1998.233333 |
| 3 | 3 | AMERICAN AIRCRAFT INC | NaN |
| 4 | 4 | AVIAT AIRCRAFT INC | 2007.000000 |
Question?
Notice the extra column named Unnamed: 0 . Where did this extra column come from?
This Unnamed: 0 column is the index from the DataFrame. Despite it not being part of the original data, it's saved with the DataFrame by default.
We can elect not to save the index with the DataFrame by passing False to the index parameter of to_csv():
avg_year_by_man_df.to_csv('../data/avg_year_by_man.csv', index=False)
And then check our result again:
pd.read_csv('../data/avg_year_by_man.csv').head()
| manufacturer | year | |
|---|---|---|
| 0 | AGUSTA SPA | 2001.000000 |
| 1 | AIRBUS | 2007.201220 |
| 2 | AIRBUS INDUSTRIE | 1998.233333 |
| 3 | AMERICAN AIRCRAFT INC | NaN |
| 4 | AVIAT AIRCRAFT INC | 2007.000000 |
The to_csv() method has similar parameters to read_csv(). A few examples:
sep- the data's delimterheader- whether or not to write out the column names
Full documentation can be pulled up by running the method name followed by a question mark:
pd.DataFrame.to_csv?
Signature: pd.DataFrame.to_csv( self, path_or_buf: 'FilePath | WriteBuffer[bytes] | WriteBuffer[str] | None' = None, sep: 'str' = ',', na_rep: 'str' = '', float_format: 'str | Callable | None' = None, columns: 'Sequence[Hashable] | None' = None, header: 'bool_t | list[str]' = True, index: 'bool_t' = True, index_label: 'IndexLabel | None' = None, mode: 'str' = 'w', encoding: 'str | None' = None, compression: 'CompressionOptions' = 'infer', quoting: 'int | None' = None, quotechar: 'str' = '"', lineterminator: 'str | None' = None, chunksize: 'int | None' = None, date_format: 'str | None' = None, doublequote: 'bool_t' = True, escapechar: 'str | None' = None, decimal: 'str' = '.', errors: 'str' = 'strict', storage_options: 'StorageOptions' = None, ) -> 'str | None' Docstring: Write object to a comma-separated values (csv) file. Parameters ---------- path_or_buf : str, path object, file-like object, or None, default None String, path object (implementing os.PathLike[str]), or file-like object implementing a write() function. If None, the result is returned as a string. If a non-binary file object is passed, it should be opened with `newline=''`, disabling universal newlines. If a binary file object is passed, `mode` might need to contain a `'b'`. .. versionchanged:: 1.2.0 Support for binary file objects was introduced. sep : str, default ',' String of length 1. Field delimiter for the output file. na_rep : str, default '' Missing data representation. float_format : str, Callable, default None Format string for floating point numbers. If a Callable is given, it takes precedence over other numeric formatting parameters, like decimal. columns : sequence, optional Columns to write. header : bool or list of str, default True Write out the column names. If a list of strings is given it is assumed to be aliases for the column names. index : bool, default True Write row names (index). index_label : str or sequence, or False, default None Column label for index column(s) if desired. If None is given, and `header` and `index` are True, then the index names are used. A sequence should be given if the object uses MultiIndex. If False do not print fields for index names. Use index_label=False for easier importing in R. mode : str, default 'w' Python write mode. The available write modes are the same as :py:func:`open`. encoding : str, optional A string representing the encoding to use in the output file, defaults to 'utf-8'. `encoding` is not supported if `path_or_buf` is a non-binary file object. compression : str or dict, default 'infer' For on-the-fly compression of the output data. If 'infer' and 'path_or_buf' is path-like, then detect compression from the following extensions: '.gz', '.bz2', '.zip', '.xz', '.zst', '.tar', '.tar.gz', '.tar.xz' or '.tar.bz2' (otherwise no compression). Set to ``None`` for no compression. Can also be a dict with key ``'method'`` set to one of {``'zip'``, ``'gzip'``, ``'bz2'``, ``'zstd'``, ``'tar'``} and other key-value pairs are forwarded to ``zipfile.ZipFile``, ``gzip.GzipFile``, ``bz2.BZ2File``, ``zstandard.ZstdCompressor`` or ``tarfile.TarFile``, respectively. As an example, the following could be passed for faster compression and to create a reproducible gzip archive: ``compression={'method': 'gzip', 'compresslevel': 1, 'mtime': 1}``. .. versionadded:: 1.5.0 Added support for `.tar` files. .. versionchanged:: 1.0.0 May now be a dict with key 'method' as compression mode and other entries as additional compression options if compression mode is 'zip'. .. versionchanged:: 1.1.0 Passing compression options as keys in dict is supported for compression modes 'gzip', 'bz2', 'zstd', and 'zip'. .. versionchanged:: 1.2.0 Compression is supported for binary file objects. .. versionchanged:: 1.2.0 Previous versions forwarded dict entries for 'gzip' to `gzip.open` instead of `gzip.GzipFile` which prevented setting `mtime`. quoting : optional constant from csv module Defaults to csv.QUOTE_MINIMAL. If you have set a `float_format` then floats are converted to strings and thus csv.QUOTE_NONNUMERIC will treat them as non-numeric. quotechar : str, default '\"' String of length 1. Character used to quote fields. lineterminator : str, optional The newline character or character sequence to use in the output file. Defaults to `os.linesep`, which depends on the OS in which this method is called ('\\n' for linux, '\\r\\n' for Windows, i.e.). .. versionchanged:: 1.5.0 Previously was line_terminator, changed for consistency with read_csv and the standard library 'csv' module. chunksize : int or None Rows to write at a time. date_format : str, default None Format string for datetime objects. doublequote : bool, default True Control quoting of `quotechar` inside a field. escapechar : str, default None String of length 1. Character used to escape `sep` and `quotechar` when appropriate. decimal : str, default '.' Character recognized as decimal separator. E.g. use ',' for European data. errors : str, default 'strict' Specifies how encoding and decoding errors are to be handled. See the errors argument for :func:`open` for a full list of options. .. versionadded:: 1.1.0 storage_options : dict, optional Extra options that make sense for a particular storage connection, e.g. host, port, username, password, etc. For HTTP(S) URLs the key-value pairs are forwarded to ``urllib.request.Request`` as header options. For other URLs (e.g. starting with "s3://", and "gcs://") the key-value pairs are forwarded to ``fsspec.open``. Please see ``fsspec`` and ``urllib`` for more details, and for more examples on storage options refer `here <https://pandas.pydata.org/docs/user_guide/io.html? highlight=storage_options#reading-writing-remote-files>`_. .. versionadded:: 1.2.0 Returns ------- None or str If path_or_buf is None, returns the resulting csv format as a string. Otherwise returns None. See Also -------- read_csv : Load a CSV file into a DataFrame. to_excel : Write DataFrame to an Excel file. Examples -------- >>> df = pd.DataFrame({'name': ['Raphael', 'Donatello'], ... 'mask': ['red', 'purple'], ... 'weapon': ['sai', 'bo staff']}) >>> df.to_csv(index=False) 'name,mask,weapon\nRaphael,red,sai\nDonatello,purple,bo staff\n' Create 'out.zip' containing 'out.csv' >>> compression_opts = dict(method='zip', ... archive_name='out.csv') # doctest: +SKIP >>> df.to_csv('out.zip', index=False, ... compression=compression_opts) # doctest: +SKIP To write a csv file to a new folder or nested folder you will first need to create it using either Pathlib or os: >>> from pathlib import Path # doctest: +SKIP >>> filepath = Path('folder/subfolder/out.csv') # doctest: +SKIP >>> filepath.parent.mkdir(parents=True, exist_ok=True) # doctest: +SKIP >>> df.to_csv(filepath) # doctest: +SKIP >>> import os # doctest: +SKIP >>> os.makedirs('folder/subfolder', exist_ok=True) # doctest: +SKIP >>> df.to_csv('folder/subfolder/out.csv') # doctest: +SKIP File: /usr/local/anaconda3/envs/uc-python/lib/python3.11/site-packages/pandas/core/generic.py Type: function
Note
There are several other df.to_xxx() methods that allow you to export DataFrames to other data formats. See more options here.
Your Turn¶
Exporting data is copying data from Python's ________ to the ________.
Fill in the blanks to the following code to:
- import the flights.csv file,
- filter for flights with a destination to the 'CVG' airport,
- write this subsetted data out to a new CSV file titled 'flights_to_cvg' (but don't save the index to the CSV).
import pandas as pd flights_df = pd.________('../data/flights.csv') flights_to_cvg_df = flights_df[flights_df[________] == 'CVG'] flights_to_cvg_df.________('../data/flights_to_cvg.csv', ________ = False)
Exporting Other Files¶
Recall being exposed to the importing of JSON and Pickle files -- now we will see how to save them.
JSON Files¶
Take a look at the below dict:
dict_example = {
"first": "Guido",
"last": "van Rossum"
}
And then we can save it as a JSON file using the with statement and the dump function from the json library:
import json
with open('../data/dict_example_export.json', 'w') as f:
f.write(json.dumps(dict_example))
We can then reimport this to verify we saved it correctly:
with open('../data/dict_example_export.json', 'r') as f:
imported_json = json.load(f)
type(imported_json)
dict
imported_json
{'first': 'Guido', 'last': 'van Rossum'}
Pickle Files¶
Question?
What are Pickle files?
Python's native data files are known as Pickle files:
- All Pickle files have the
.pickleextension
- Pickle files are great for saving native Python data that can't easily be represented by other file types
- Pre-processed data
- Models
- Any other Python object...
Exporting Pickle Files¶
Pickle files can be exported using the pickle library paired with the with statement and the open() function:
import pickle
with open('../data/pickle_example_export.pickle', 'wb') as f:
pickle.dump(dict_example, f)
We can then reimport this to verify we saved it correctly:
with open('../data/pickle_example_export.pickle', 'rb') as f:
imported_pickle = pickle.load(f)
type(imported_pickle)
dict
imported_pickle
{'first': 'Guido', 'last': 'van Rossum'}
Questions¶
Are there any questions before we move on?