Modular Code¶

What's Modularity?¶
- Building our code in discrete, clearly separated chunks
- So we can...
- Modify one piece without breaking the others
- Combine pieces in different ways, as we need them
Often, modularity implies breaking our code down into different functions which live in different modules.
Note
Generally, a module in Python is just a file that ends in .py
How do we achieve modularity in Python?¶
- Move code chunks into their own functions
- Move functions into their own files
Example: Functions¶
In the last section, we imported some data from a CSV, kept just its numeric columns, and separated the target from the features...
import numpy as np
import pandas as pd
adult_census = pd.read_csv("../data/adult-census.csv")
# create column names of interest
target_col = "class"
raw_features = adult_census.drop(columns=target_col)
numeric_features = raw_features.select_dtypes(np.number)
feature_cols = numeric_features.columns.values
features = adult_census[feature_cols]
target = adult_census[target_col]
This is a lot of code though, and we might want to do this again in the future with different data sets.
That makes it a perfect case to be its own function.
def get_features_and_target():
'''Split a CSV into a DF of numeric features and a target column.'''
adult_census = pd.read_csv("../data/adult-census.csv")
target_col = "class"
raw_features = adult_census.drop(columns=target_col)
numeric_features = raw_features.select_dtypes(np.number)
feature_cols = numeric_features.columns.values
features = adult_census[feature_cols]
target = adult_census[target_col]
return (features, target)
Let's test it!
f, t = get_features_and_target()
f.head()
| age | education-num | capital-gain | capital-loss | hours-per-week | |
|---|---|---|---|---|---|
| 0 | 25 | 7 | 0 | 0 | 40 |
| 1 | 38 | 9 | 0 | 0 | 50 |
| 2 | 28 | 12 | 0 | 0 | 40 |
| 3 | 44 | 10 | 7688 | 0 | 40 |
| 4 | 18 | 10 | 0 | 0 | 30 |
t.head()
0 <=50K 1 <=50K 2 >50K 3 >50K 4 <=50K Name: class, dtype: object
Looks like it worked!!
def get_features_and_target():
'''Split a CSV into a DF of numeric features and a target column.'''
adult_census = pd.read_csv("../data/adult-census.csv")
target_col = "class"
raw_features = adult_census.drop(columns=target_col)
numeric_features = raw_features.select_dtypes(np.number)
feature_cols = numeric_features.columns.values
features = adult_census[feature_cols]
target = adult_census[target_col]
return (features, target)
Discussion
How would we apply this function to new data?We can't! We didn't parametrize it.
Parametrizing Functions¶
While functions are about reusing code, we rarely want to rerun exactly the same code.
Usually, there are a small number of things that should change from run to run. These are called parameters.
Common things that might be used parameters:
- threshold values
- filenames
- column names
def get_features_and_target():
'''Split a CSV into a DF of numeric features and a target column.'''
adult_census = pd.read_csv("../data/adult-census.csv")
target_col = "class"
raw_features = adult_census.drop(columns=target_col)
numeric_features = raw_features.select_dtypes(np.number)
feature_cols = numeric_features.columns.values
features = adult_census[feature_cols]
target = adult_census[target_col]
return (features, target)
Discussion
What should be the parameters of ourget_features_and_target function?
def get_features_and_target(csv_file, target_col):
'''Split a CSV into a DF of numeric features and a target column.'''
adult_census = pd.read_csv(csv_file)
raw_features = adult_census.drop(columns=target_col)
numeric_features = raw_features.select_dtypes(np.number)
feature_cols = numeric_features.columns.values
features = adult_census[feature_cols]
target = adult_census[target_col]
return (features, target)
Now if we call our function without passing csv_file and target_col, we get an error:
f, t = get_features_and_target()
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) /var/folders/j3/v1318ng94fvdpq7kzr0hq9kw0000gn/T/ipykernel_2533/3218846325.py in <module> ----> 1 f, t = get_features_and_target() TypeError: get_features_and_target() missing 2 required positional arguments: 'csv_file' and 'target_col'
But we can still use it if we pass in those parameters:
# In Python, linebreaks and spaces inside parentheses are ignored.
f, t = get_features_and_target(
csv_file='../data/adult-census.csv',
target_col='class',
)
f.head()
| age | education-num | capital-gain | capital-loss | hours-per-week | |
|---|---|---|---|---|---|
| 0 | 25 | 7 | 0 | 0 | 40 |
| 1 | 38 | 9 | 0 | 0 | 50 |
| 2 | 28 | 12 | 0 | 0 | 40 |
| 3 | 44 | 10 | 7688 | 0 | 40 |
| 4 | 18 | 10 | 0 | 0 | 30 |
Now, imagine we want to build a model on the Ames data instead, using the "Sale_Price" column as our target...
ames_features, ames_target = get_features_and_target(
csv_file='../data/ames.csv',
target_col='Sale_Price',
)
ames_features.head()
| Lot_Frontage | Lot_Area | Year_Built | Year_Remod_Add | Mas_Vnr_Area | BsmtFin_SF_1 | BsmtFin_SF_2 | Bsmt_Unf_SF | Total_Bsmt_SF | First_Flr_SF | ... | Open_Porch_SF | Enclosed_Porch | Three_season_porch | Screen_Porch | Pool_Area | Misc_Val | Mo_Sold | Year_Sold | Longitude | Latitude | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 141 | 31770 | 1960 | 1960 | 112 | 2 | 0 | 441 | 1080 | 1656 | ... | 62 | 0 | 0 | 0 | 0 | 0 | 5 | 2010 | -93.619754 | 42.054035 |
| 1 | 80 | 11622 | 1961 | 1961 | 0 | 6 | 144 | 270 | 882 | 896 | ... | 0 | 0 | 0 | 120 | 0 | 0 | 6 | 2010 | -93.619756 | 42.053014 |
| 2 | 81 | 14267 | 1958 | 1958 | 108 | 1 | 0 | 406 | 1329 | 1329 | ... | 36 | 0 | 0 | 0 | 0 | 12500 | 6 | 2010 | -93.619387 | 42.052659 |
| 3 | 93 | 11160 | 1968 | 1968 | 0 | 1 | 0 | 1045 | 2110 | 2110 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2010 | -93.617320 | 42.051245 |
| 4 | 74 | 13830 | 1997 | 1998 | 0 | 3 | 0 | 137 | 928 | 928 | ... | 34 | 0 | 0 | 0 | 0 | 0 | 3 | 2010 | -93.638933 | 42.060899 |
5 rows × 34 columns
We've successfully abstracted some of our code logic, moving it to a separate function that we can use without having to think too much about how it works.
This is the foundation of building larger projects in Python.
Example: Files¶
As we write more and more functions, it can be nice to move them outside of the script or notebook where we're currently working.
Let's move our new function into its own file, or module, and then use it from Jupyter.
We'll start by creating a new text file in Jupyter:
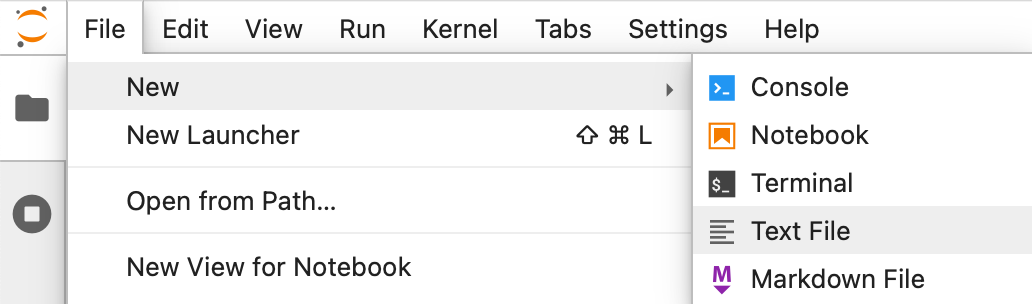
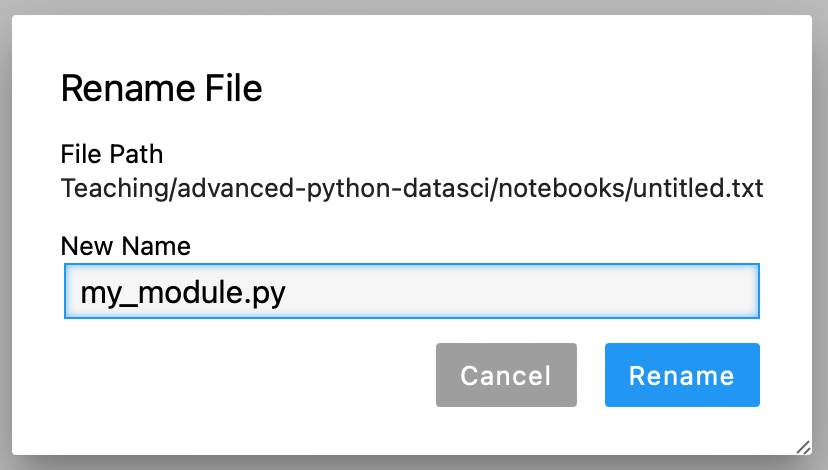
Then we'll give our new module a sensible name. Right-click on the untitled.txt tab and rename it to my_module.py.
Don't forget to make sure it ends in .py, not .txt!
Then paste the function we wrote, along with lines to import numpy and pandas:
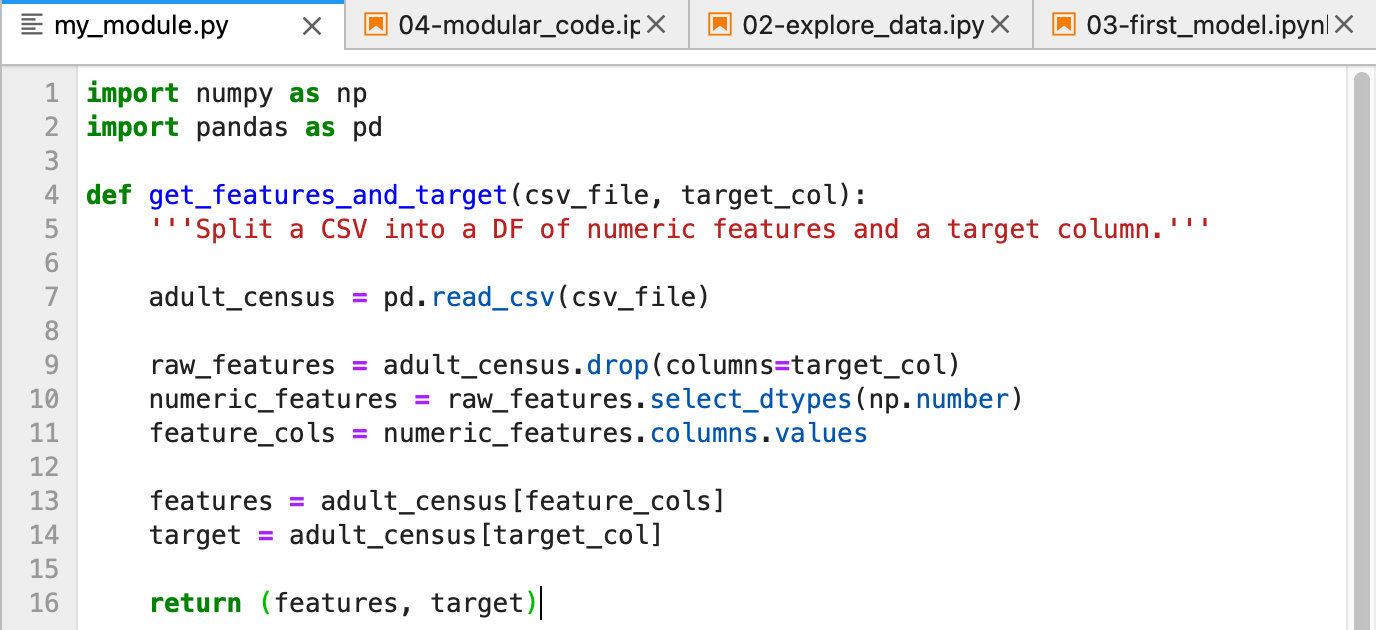
Save the file and close the my_module.py tab.
Notice how that file is now in your sidebar:
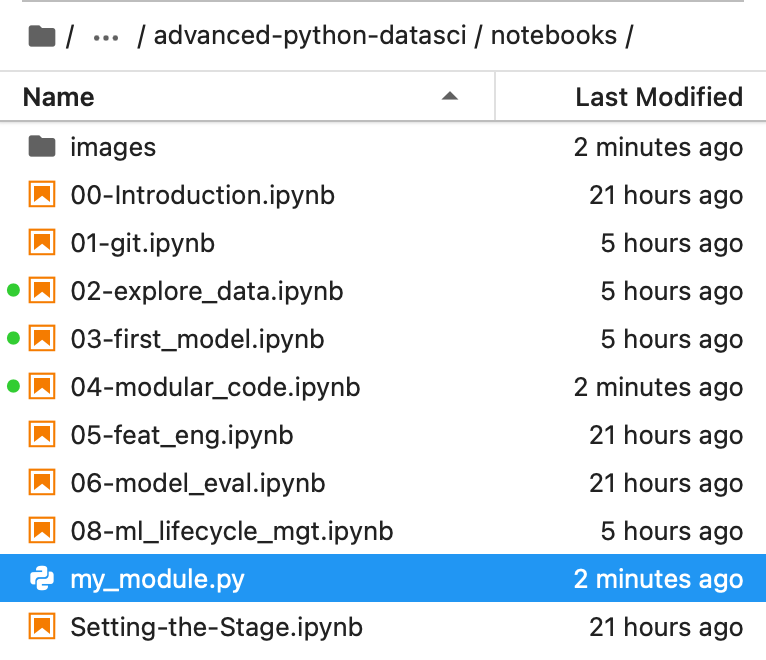
Now that our module is saved, we can import it in any notebook (or script) that's saved in the same folder as the module.
import my_module
my_module.get_features_and_target?
Signature: my_module.get_features_and_target(csv_file, target_col) Docstring: Split a CSV into a DF of numeric features and a target column. File: ~/Teaching/advanced-python-datasci/notebooks/my_module.py Type: function
f, t = my_module.get_features_and_target(
csv_file='../data/adult-census.csv',
target_col='class',
)
f.head()
| age | education-num | capital-gain | capital-loss | hours-per-week | |
|---|---|---|---|---|---|
| 0 | 25 | 7 | 0 | 0 | 40 |
| 1 | 38 | 9 | 0 | 0 | 50 |
| 2 | 28 | 12 | 0 | 0 | 40 |
| 3 | 44 | 10 | 7688 | 0 | 40 |
| 4 | 18 | 10 | 0 | 0 | 30 |
Notice that we call our function as my_module.get_features_and_target, not just get_features_and_target.
Discussion
Does this syntax remind you of anything we've seen before?Ultimately, modules you create aren't any different from numpy, pandas, or any other Python libraries. You can build them and use them just the same way.
It's a good idea to put related functions into a module, which you can then reuse within a project or even across different projects.
Committing to GitHub¶
Before we end this section, let's commit our code so far to GitHub.
Open GitHub Desktop. It should show you what files you've added and changed in the project. In the summary box, write a message that encapsulates what we've done so far.
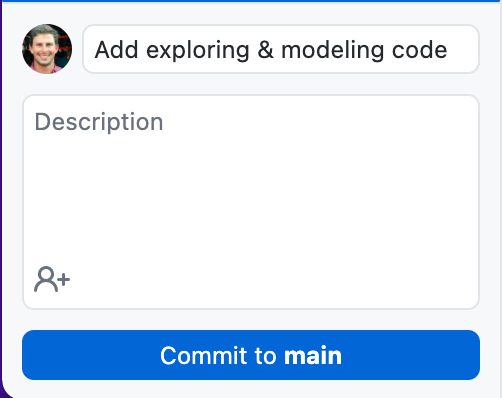
Then press "Commit to main".
At this point, we've committed our code but haven't synced it with GitHub. GitHub Desktop will inform us that we need to "push" our new commit:
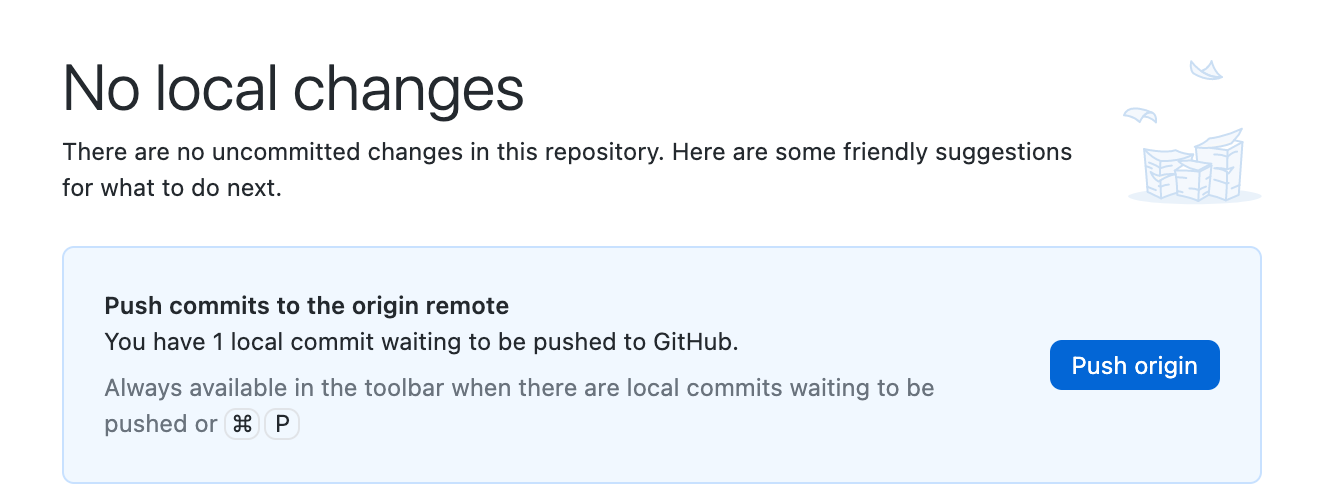
Click the "Push Origin" button. Done!
If you go to your advanced-python-datasci repo in GitHub, you should see your new files!
Questions¶
Are there any questions before we move on?